RNA Center Software
Pymol plugins for axis and angle rotation
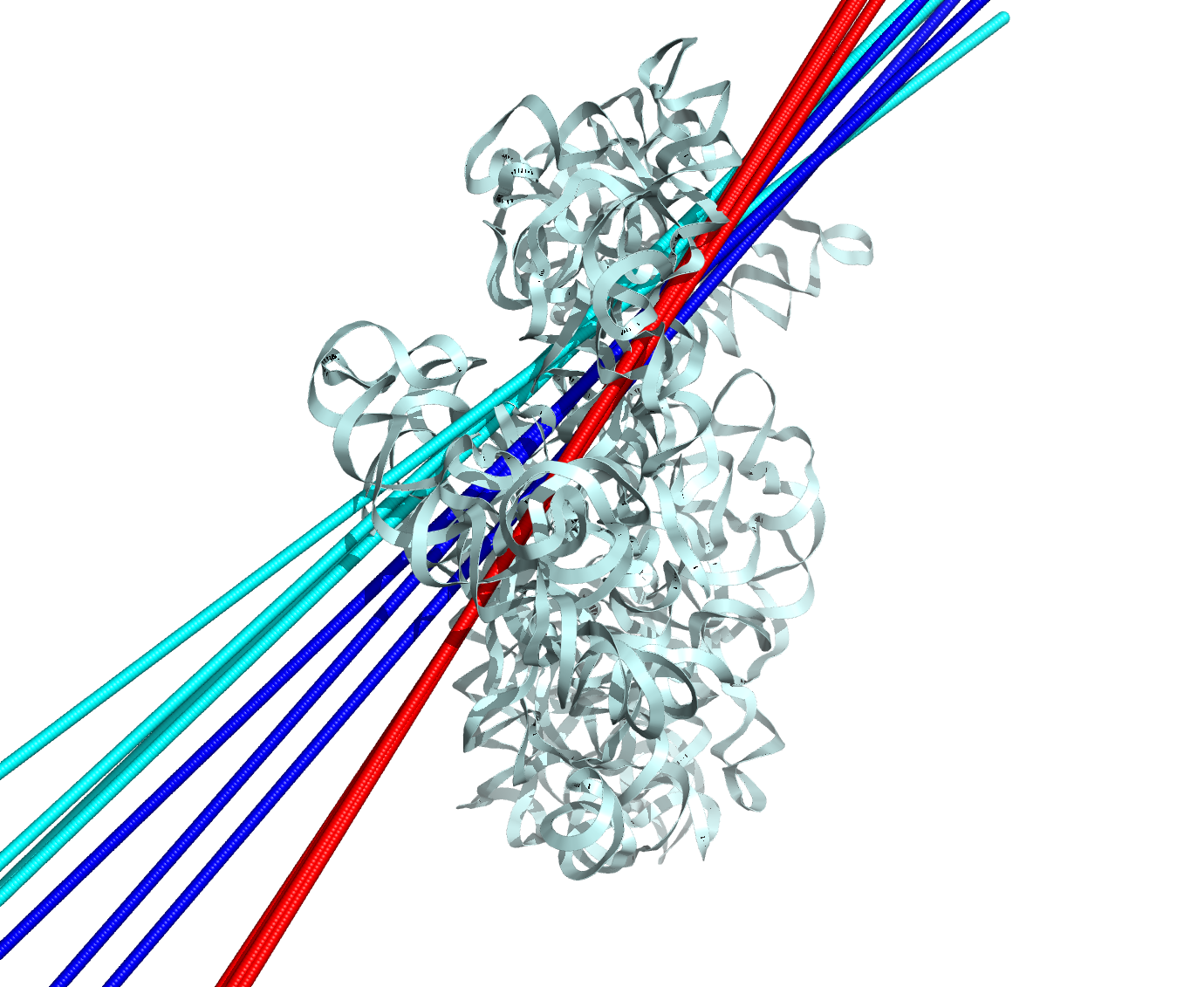
- Pymol plugin for Euler-Rodriguez Transformation calculation of axis and angle or rotation (…save as)
- If you use this plug-in, please cite: Mohan, S., Donohue, J. P. & Noller, H. F. Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. U. S. A. 111, 13325-13330.
Math and matrices
The molecular region of interest for the reference structure is held fixed.The comparison molecular region is aligned to the reference.
The 4x4 matrix of this transformation matrix is obtained from pyMOL.
The matrix is a combination of a rotation and translation.
Eulers rotation theorem states that any rotation can be expressed as a single rotation about some axis.
An axis and rotation angle is obtained such that the motion is equivalent to one rotation.
Xa is original coordinates, Xb is coordinates in aligned to the reference structure.
Let R be the rotation matrix , T is the translation, O can be thought of as an origin shift.
Xb = R Xa + T
Xb + O = R (Xa + O)
R Xa + T = R (Xa + O) - O
R Xa + T = R Xa + R O - O
T = R O - O
T = (R - I) O
O = inv(R-I) T
Technically a pseudo-inverse is used which is equivalent to a 'best fit' (least squares) solution
The angle of rotation is derived from the Euler-Rodrigues formula.
theta = acos((R[0][0] + R[1][1] + R[2][2] -1)/2)
The direction of the axis is similarly calculated.
e0 = (R[2][1] - R[1][2]) / (2*sin(theta))
e1 = (R[0][2] - R[2][0]) / (2*sin(theta))
e2 = (R[1][0] - R[0][1]) / (2*sin(theta))
The axis can now be placed in the e direction passing through the point O.
pymol syntax
load 2J00.pdb.gz
run erodaxis.py
load ../1GIX.pdb.gz
align 1GIX and chain A and not resi 930:1390 , 2J00 and chain A and not resi 930:1390
create yushead , 2J00 and chain A and resi 930:1390
create otherhead , 1GIX and chain A and resi 930:1390
erodaxis otherhead , yushead